Show code cell source
import numpy as np
import scipy as sp
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
import matplotlib as mp
import sklearn
import networkx as nx
from IPython.display import Image, HTML
import laUtilities as ut
%matplotlib inline
Gradient Descent#
Most of the machine learning we have studied this semester is based on the idea that we have a model that is parameterized, and our goal is to find good settings for the parameters.
We have seen example after example of this problem.
In \(k\)-means, our goal was to find \(k\) cluster centroids, so that the \(k\)-means objective was minimized.
In linear regression, our goal was to find a parameter vector \(\beta\) so that sum of squared error \(\Vert \mathbf{y} - \hat{\mathbf{y}}\Vert\) was minimized.
In the support vector machine, our goal was to find a parameter vector \(\theta\) so that classification error was minimized.
And on and on …
It’s time now to talk about how, in general, one can find “good settings” for the parameters in problems like these.
What allows us to unify our approach to many such problems is the following:
First, we start by defining an error function, generally called a loss function, to describe how well our method is doing.
And second, we choose loss functions that are differentiable with respect to the parameters.
These two requirements mean that we can think of the parameter tuning problem using surfaces like these:
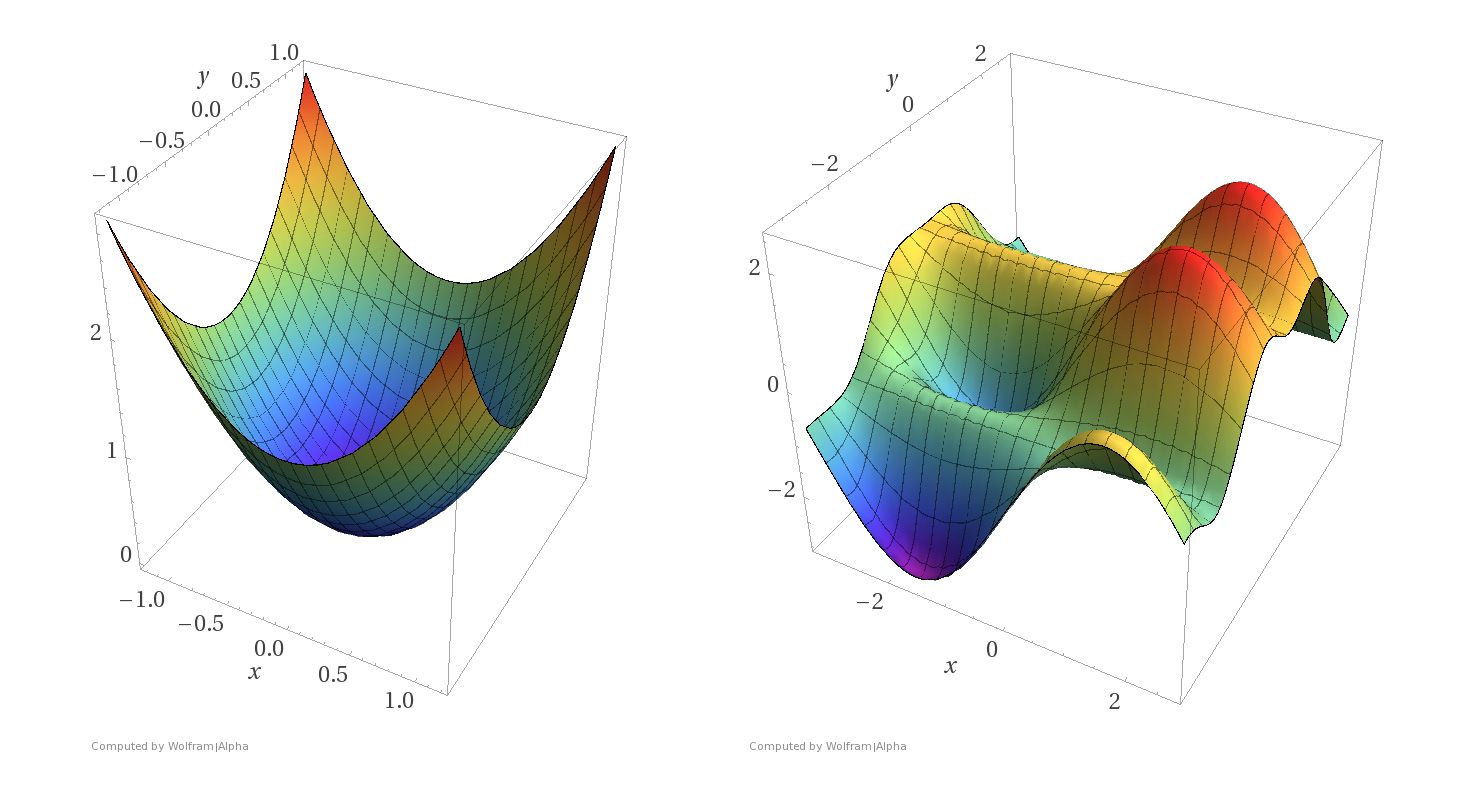
Imagine that the \(x\) and \(y\) axes in these pictures represent parameter settings. That is, we have two parameters to set, corresponding to the values of \(x\) and \(y\).
For each \((x, y)\) setting, the \(z\)-axis shows the value of the loss function.
What we want to do is find the minimum of a surface, corresponding to the parameter settings that minimize loss.
Notice the difference between the two kinds of surfaces.
The surface on the left corresponds to a strictly convex loss function. If we find a local minimum of this function, it is a global minimum.
The surface on the right corresponds to a non-convex loss function. There are local minima that are not globally minimal.
Both kinds of loss functions arise in machine learning.
For example, convex loss functions arise in
Linear regression
Logistic regression
While non-convex loss functions arise in
\(k\)-means
Gaussian Mixture Modeling
and many other settings
Gradient Descent Intuitively#
The intuition of gradient descent is the following.
Imagine you are lost in the mountains, and it is foggy out. You want to find a valley. But since it is foggy, you can only see the local area around you.

The natural thing to do is:
Look around you 360 degrees.
Observe in which direction the ground is sloping downward most steeply.
Take a few steps in that direction.
Repeat the process
… until the ground seems to be level.
Formalizing Gradient Descent#
The key to this intuitive idea is formalizing the idea of “direction of steepest descent.”
This is where the differentiability of the loss function comes into play.
As long as the loss function is differentiable, we can define the direction of steepest descent (really, ascent).
That direction is called the gradient.
The gradient is a generalization of the slope of a line.
Let’s say we have a loss function \(\mathcal{L}(\mathbf{w})\).
The components of \(\mathbf{w}\in\mathbb{R}^n\) are the parameters we want to optimize.
For example, if our problem is linear regression, the loss function could be:
where \(\hat{\mathbf{y}}\) is our estimate, ie, \(\hat{\mathbf{y}} = X\mathbf{w}\) so that
To find the gradient, we take the partial derivative of our loss function with respect to each parameter:
and collect all the partial derivatives into a vector of the same shape as \(\mathbf{w}\):
When you see the notation \(\nabla_\mathbf{w}\mathcal{L},\) think of it as \( \frac{d\mathcal{L}}{d\mathbf{w}} \), keeping in mind that \(\mathbf{w}\) is a vector.
It turns out that if we are going to take a small step of unit length, then the gradient is the direction that maximizes the change in the loss function.
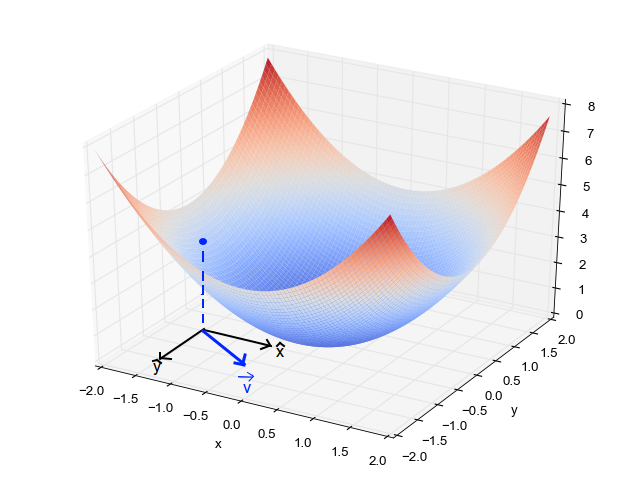
As you can see from the above figure, in general the gradient varies depending on where you are in the parameter space.
So we write:
Each time we seek to improve our parameter estimates \(\mathbf{w}\), we will take a step in the negative direction of the gradient.
… “negative direction” because the gradient specifies the direction of maximum increase – and we want to decrease the loss function.
How big a step should we take?
For step size, will use a scalar value \(\eta\) called the learning rate.
The learning rate is a hyperparameter that needs to be tuned for a given problem, or even can be modified adaptively as the algorithm progresses.
Now we can write the gradient descent algorithm formally:
Start with an initial parameter estimate \(\mathbf{w}^0\).
Update: \(\mathbf{w}^{n+1} = \mathbf{w}^n - \eta \nabla_\mathbf{w}\mathcal{L}(\mathbf{w}^n)\)
If not converged, go to step 2.
How do we know if we are “converged”?
Typically we stop the iteration if the loss has not improved by a fixed amount for a pre-decided number, say 10 or 20, iterations.
Example: Linear Regression#
Let’s say we have this dataset.
Show code cell source
def centerAxes(ax):
ax.spines['left'].set_position('zero')
ax.spines['right'].set_color('none')
ax.spines['bottom'].set_position('zero')
ax.spines['top'].set_color('none')
ax.xaxis.set_ticks_position('bottom')
ax.yaxis.set_ticks_position('left')
bounds = np.array([ax.axes.get_xlim(), ax.axes.get_ylim()])
ax.plot(bounds[0][0],bounds[1][0],'')
ax.plot(bounds[0][1],bounds[1][1],'')
n = 10
beta = np.array([1., 0.5])
ax = plt.figure(figsize = (7, 7)).add_subplot()
centerAxes(ax)
np.random.seed(1)
xlin = -10.0 + 20.0 * np.random.random(n)
y = beta[0] + (beta[1] * xlin) + np.random.randn(n)
ax.plot(xlin, y, 'ro', markersize = 10);
Let’s fit a least-squares line to this data.
The loss function for this problem is the least-squares error:
Of course, we know how to solve this problem using the normal equations, but let’s do it using gradient descent instead.
Here is the line we’d like to find:
Show code cell source
ax = plt.figure(figsize = (7, 7)).add_subplot()
centerAxes(ax)
ax.plot(xlin, y, 'ro', markersize = 10)
ax.plot(xlin, beta[0] + beta[1] * xlin, 'b-')
plt.text(-9, 3, r'$y = \beta_0 + \beta_1x$', size=20);
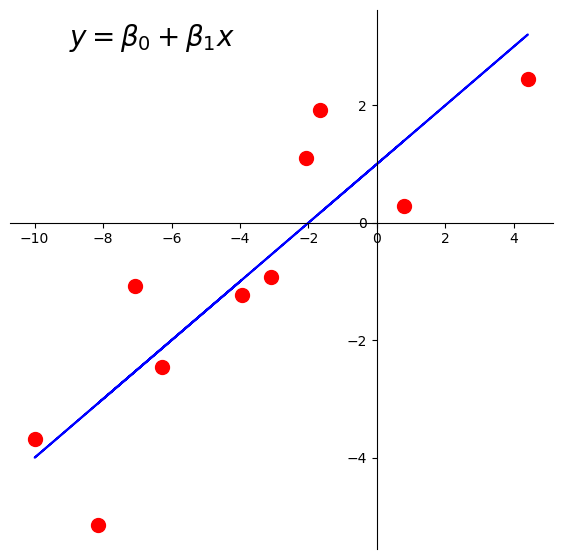
There are \(n = 10\) data points, whose \(x\) and \(y\) values are stored in xlin and y.
First, let’s create our \(X\) (design) matrix, and include a column of ones to model the intercept:
X = np.column_stack([np.ones((n, 1)), xlin])
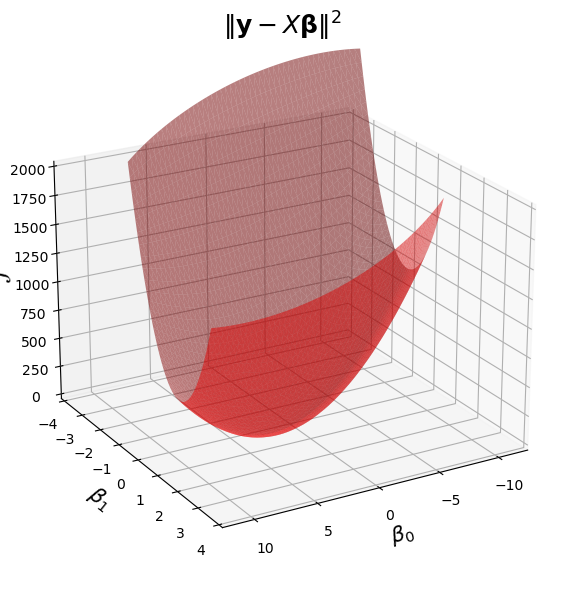
Now, let’s visualize the loss function \(\mathcal{L}(\mathbf{\beta}) = \Vert \mathbf{y}-X\mathbf{\beta}\Vert^2.\)
Show code cell source
fig = ut.three_d_figure((23, 1), '',
-12, 12, -4, 4, -1, 2000,
figsize = (7, 7))
qf = np.array(X.T @ X)
fig.ax.view_init(azim = 60, elev = 22)
fig.plotGeneralQF(X.T @ X, -2 * (y.T @ X), y.T @ y, alpha = 0.5)
fig.ax.set_zlabel('$\mathcal{L}$')
fig.ax.set_xlabel(r'$\beta_0$')
fig.ax.set_ylabel(r'$\beta_1$')
fig.set_title(r'$\Vert \mathbf{y}-X\mathbf{\beta}\Vert^2$', '',
number_fig = False, size = 18)
# fig.save();
I won’t take you through computing the gradient for this problem (you can find it in the online text).
I will just tell you that the gradient for a least squares problem is:
Note
For those interested in a little more insight into what these plots are showing, here is the derivation.
We start from the rule that \(\Vert \mathbf{v}\Vert = \sqrt{\mathbf{v}^T\mathbf{v}}\).
Applying this rule to our loss function:
The first term, \(\beta^T X^T X \beta\), is a quadratic form, and it is what makes this surface curved. As long as \(X\) has independent columns, \(X^TX\) is positive definite, so the overall shape is a paraboloid opening upward, and the surface has a unique minimum point.
To find the gradient, we can use standard calculus rules for derivates involving vectors. The rules are not complicated, but the bottom line is that in this case, you can almost use the same rules you would if \(\beta\) were a scalar:
And by the way – since we’ve computed the derivative as a function of \(\beta\), instead of using gradient descent, we could simply solve for the point where the gradient is zero. This is the optimal point which we know must exist:
Which of course, are the normal equations for this linear system.
So here is our code for gradient descent:
def loss(X, y, beta):
return np.linalg.norm(y - X @ beta) ** 2
def gradient(X, y, beta):
return X.T @ X @ beta - X.T @ y
def gradient_descent(X, y, beta_hat, eta, nsteps = 1000):
losses = [loss(X, y, beta_hat)]
betas = [beta_hat]
#
for step in range(nsteps):
#
# the gradient step
new_beta_hat = beta_hat - eta * gradient(X, y, beta_hat)
beta_hat = new_beta_hat
#
# accumulate statistics
losses.append(loss(X, y, new_beta_hat))
betas.append(new_beta_hat)
return np.array(betas), np.array(losses)
We’ll start at an arbitrary point, say, \((-8, -3.2)\).
That is, \(\beta_0 = -8\), and \(\beta_1 = -3.2\).
beta_start = np.array([-8, -3.2])
eta = 0.002
betas, losses = gradient_descent(X, y, beta_start, eta)
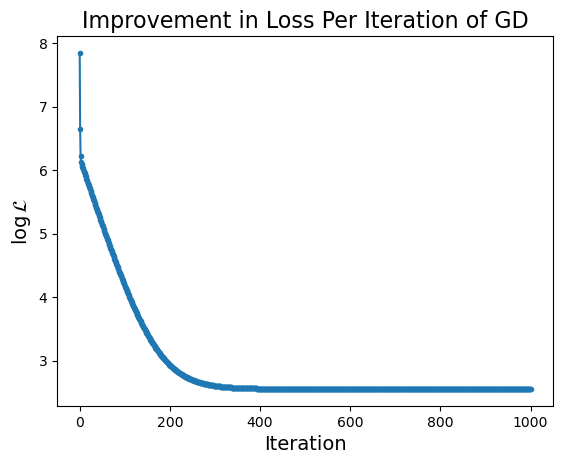
What happens to our loss function per GD iteration?
Show code cell source
plt.plot(np.log(losses), '.-')
plt.ylabel(r'$\log\mathcal{L}$', size = 14)
plt.xlabel('Iteration', size = 14)
plt.title('Improvement in Loss Per Iteration of GD', size = 16);
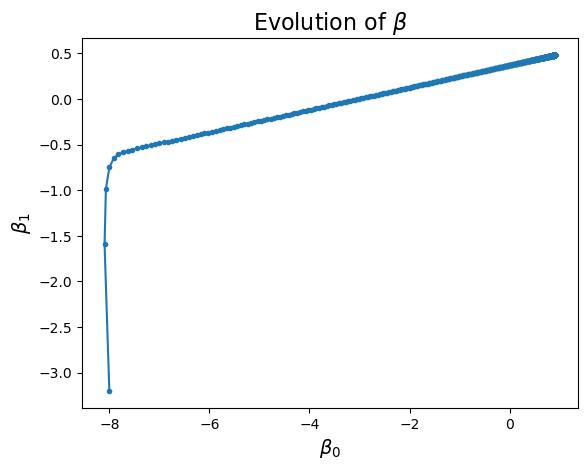
Show code cell source
plt.plot(betas[:, 0], betas[:, 1], '.-')
plt.xlabel(r'$\beta_0$', size = 14)
plt.ylabel(r'$\beta_1$', size = 14)
plt.title(r'Evolution of $\beta$', size = 16);
Notice that the improvement in loss decreases over time. Initially the gradient is steep and loss improves fast, while later on the gradient is shallow and loss doesn’t improve much per step.
Now remember that in reality we are like the person who is trying to find their way down the mountain, in the fog.
In general we cannot “see” the entire loss function surface.
Nonetheless, since we know what the loss surface looks like in this case, we can visualize the algorithm “moving” on that surface.
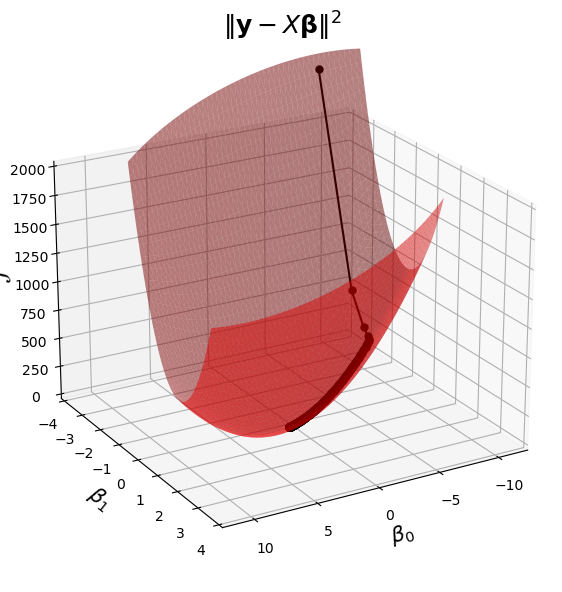
This visualization combines the last two plots into a single view.
Show code cell source
%matplotlib inline
fig = ut.three_d_figure((23, 1), '',
-12, 12, -4, 4, -1, 2000,
figsize = (7, 7))
qf = np.array(X.T @ X)
fig.ax.view_init(azim = 60, elev = 22)
fig.plotGeneralQF(X.T @ X, -2 * (y.T @ X), y.T @ y, alpha = 0.5)
fig.ax.set_zlabel('$\mathcal{L}$')
fig.ax.set_xlabel(r'$\beta_0$')
fig.ax.set_ylabel(r'$\beta_1$')
fig.set_title(r'$\Vert \mathbf{y}-X\mathbf{\beta}\Vert^2$', '',
number_fig = False, size = 18)
fig.ax.plot(betas[:, 0], betas[:, 1], 'o-', zs = losses, c = 'k', markersize = 5);
#
Next we can visualize how the algorithm progresses, both in parameter space and in data space:
Show code cell source
%matplotlib inline
# set up view
import matplotlib.animation as animation
mp.rcParams['animation.html'] = 'jshtml'
anim_frames = np.array(list(range(10)) + [2 * x for x in range(5, 25)] + [5 * x for x in range(10, 100)])
fig = ut.three_d_figure((23, 1), 'z = 3 x1^2 + 7 x2 ^2',
-12, 12, -4, 4, -1, 2000,
figsize = (7, 7))
plt.close()
fig.ax.view_init(azim = 60, elev = 22)
qf = np.array(X.T @ X)
fig.plotGeneralQF(X.T @ X, -2 * (y.T @ X), y.T @ y, alpha = 0.5)
fig.ax.set_zlabel('$\mathcal{L}$')
fig.ax.set_xlabel(r'$\beta_0$')
fig.ax.set_ylabel(r'$\beta_1$')
fig.set_title(r'$\Vert \mathbf{y}-X\mathbf{\beta}\Vert^2$', '',
number_fig = False, size = 18)
#
def anim(frame):
fig.ax.plot(betas[:frame, 0], betas[:frame, 1], 'o-', zs = losses[:frame], c = 'k', markersize = 5)
# fig.canvas.draw()
#
# create the animation
animation.FuncAnimation(fig.fig, anim,
frames = anim_frames,
fargs = None,
interval = 1,
repeat = False)
Animation size has reached 20999994 bytes, exceeding the limit of 20971520.0. If you're sure you want a larger animation embedded, set the animation.embed_limit rc parameter to a larger value (in MB). This and further frames will be dropped.